class: title-slide <br> <br> .right-panel[ # Changing Variables ## Dr. Mine Dogucu ] --- ``` r glimpse(arthritis) ``` ``` ## Rows: 530 ## Columns: 5 ## $ age <dbl> 85, 86, 83, 83, 85, 79, 90, 90, 87, 82, 77, 86, 84, 76, 77… ## $ age_gp <fct> elderly, elderly, elderly, elderly, elderly, elderly, elde… ## $ sex <fct> female, female, female, female, female, male, female, fema… ## $ yrs_from_dx <dbl> 27, 27, 10, 9, NA, NA, 51, 11, 36, 4, 31, NA, 9, 10, 3, 10… ## $ cdai <dbl> NA, 23.0, 14.5, NA, NA, NA, NA, 40.0, 6.0, NA, 0.0, NA, NA… ``` **Goal**: Create a new variable called `age_months` that represents `age` in months. --- ``` r arthritis %>% mutate(age_months = age*12) ``` ``` ## # A tibble: 530 × 6 ## age age_gp sex yrs_from_dx cdai age_months ## <dbl> <fct> <fct> <dbl> <dbl> <dbl> ## 1 85 elderly female 27 NA 1020 ## 2 86 elderly female 27 23 1032 ## 3 83 elderly female 10 14.5 996 ## 4 83 elderly female 9 NA 996 ## 5 85 elderly female NA NA 1020 ## 6 79 elderly male NA NA 948 ## 7 90 elderly female 51 NA 1080 ## 8 90 elderly female 11 40 1080 ## 9 87 elderly female 36 6 1044 ## 10 82 elderly female 4 NA 984 ## # ℹ 520 more rows ``` --- ``` r glimpse(arthritis) ``` ``` ## Rows: 530 ## Columns: 5 ## $ age <dbl> 85, 86, 83, 83, 85, 79, 90, 90, 87, 82, 77, 86, 84, 76, 77… ## $ age_gp <fct> elderly, elderly, elderly, elderly, elderly, elderly, elde… ## $ sex <fct> female, female, female, female, female, male, female, fema… ## $ yrs_from_dx <dbl> 27, 27, 10, 9, NA, NA, 51, 11, 36, 4, 31, NA, 9, 10, 3, 10… ## $ cdai <dbl> NA, 23.0, 14.5, NA, NA, NA, NA, 40.0, 6.0, NA, 0.0, NA, NA… ``` --- ``` r arthritis_age_mon <- arthritis %>% mutate(age_months = age*12) glimpse(arthritis_age_mon) ``` ``` ## Rows: 530 ## Columns: 6 ## $ age <dbl> 85, 86, 83, 83, 85, 79, 90, 90, 87, 82, 77, 86, 84, 76, 77… ## $ age_gp <fct> elderly, elderly, elderly, elderly, elderly, elderly, elde… ## $ sex <fct> female, female, female, female, female, male, female, fema… ## $ yrs_from_dx <dbl> 27, 27, 10, 9, NA, NA, 51, 11, 36, 4, 31, NA, 9, 10, 3, 10… ## $ cdai <dbl> NA, 23.0, 14.5, NA, NA, NA, NA, 40.0, 6.0, NA, 0.0, NA, NA… ## $ age_months <dbl> 1020, 1032, 996, 996, 1020, 948, 1080, 1080, 1044, 984, 92… ``` --- **Goal**: Create a new variable called `cdai_level` which reflects the levels in the the data dictionary - Remission ≤ 2.8 - Low Disease Activity > 2.8 and ≤ 10 - Moderate Disease Activity > 10 and ≤ 22 - High Disease Activity > 22 --- ``` r arthritis %>% mutate(cdai_level = case_when( cdai <=2.8 ~ "Remission", cdai > 2.8 & cdai <= 10 ~ "Low", cdai > 10 & cdai <=22 ~ "Moderate", cdai > 22 ~ "High")) ``` ``` ## # A tibble: 530 × 6 ## age age_gp sex yrs_from_dx cdai cdai_level ## <dbl> <fct> <fct> <dbl> <dbl> <chr> ## 1 85 elderly female 27 NA <NA> ## 2 86 elderly female 27 23 High ## 3 83 elderly female 10 14.5 Moderate ## 4 83 elderly female 9 NA <NA> ## 5 85 elderly female NA NA <NA> ## 6 79 elderly male NA NA <NA> ## 7 90 elderly female 51 NA <NA> ## 8 90 elderly female 11 40 High ## 9 87 elderly female 36 6 Low ## 10 82 elderly female 4 NA <NA> ## # ℹ 520 more rows ``` --- We can't really see what we have created --- We can use pipes with ggplot too! .left-panel[ ``` r arthritis %>% mutate(cdai_level = case_when( cdai <=2.8 ~ "Remission", cdai > 2.8 & cdai <= 10 ~ "Low", cdai > 10 & cdai <=22 ~ "Moderate", cdai > 22 ~ "High")) %>% ggplot(aes(x = cdai_level)) + geom_bar() ``` ] .right-panel[ 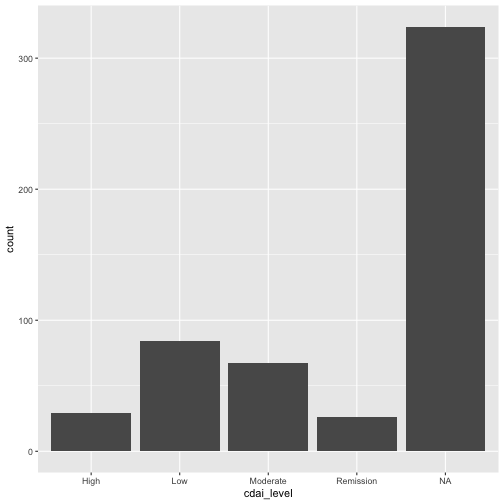<!-- --> ] --- ``` r arthritis <- arthritis %>% mutate(cdai_level = case_when( cdai <=2.8 ~ "Remission", cdai > 2.8 & cdai <= 10 ~ "Low", cdai > 10 & cdai <=22 ~ "Moderate", cdai > 22 ~ "High")) ``` --- ``` r glimpse(arthritis) ``` ``` ## Rows: 530 ## Columns: 6 ## $ age <dbl> 85, 86, 83, 83, 85, 79, 90, 90, 87, 82, 77, 86, 84, 76, 77… ## $ age_gp <fct> elderly, elderly, elderly, elderly, elderly, elderly, elde… ## $ sex <fct> female, female, female, female, female, male, female, fema… ## $ yrs_from_dx <dbl> 27, 27, 10, 9, NA, NA, 51, 11, 36, 4, 31, NA, 9, 10, 3, 10… ## $ cdai <dbl> NA, 23.0, 14.5, NA, NA, NA, NA, 40.0, 6.0, NA, 0.0, NA, NA… ## $ cdai_level <chr> NA, "High", "Moderate", NA, NA, NA, NA, "High", "Low", NA,… ``` **Goal**: Make `cdai_level` a factor and `age` an integer. --- ``` r arthritis %>% mutate(cdai_level = as.factor(cdai_level), age = as.integer(age)) ``` ``` ## # A tibble: 530 × 6 ## age age_gp sex yrs_from_dx cdai cdai_level ## <int> <fct> <fct> <dbl> <dbl> <fct> ## 1 85 elderly female 27 NA <NA> ## 2 86 elderly female 27 23 High ## 3 83 elderly female 10 14.5 Moderate ## 4 83 elderly female 9 NA <NA> ## 5 85 elderly female NA NA <NA> ## 6 79 elderly male NA NA <NA> ## 7 90 elderly female 51 NA <NA> ## 8 90 elderly female 11 40 High ## 9 87 elderly female 36 6 Low ## 10 82 elderly female 4 NA <NA> ## # ℹ 520 more rows ``` --- class: middle `as.factor()` - makes a vector factor `as.numeric()` - makes a vector numeric `as.integer()` - makes a vector integer `as.double()` - makes a vector double `as.character()` - makes a vector character